
ResearchResearch
1.A.1 Photo-reaction of Photoactive Yellow Protein (PYP)
@@PYP is a protein isolated from the purple sulfur bacterium Ectothiorhodospira
halophila. It is considered to possess a function of a blue light photoreceptor
for a negative phototactic response. It is a relatively small (14 KD) water
soluble protein. The chromophore of PYP is p-coumaric acid (4-hydroxycinnamic
acid) covalently bound to the side chain of Cys 69 via a thioester linkage.
After photoexcitation, PYP undergoes a complete photocycle triggered by
the photoisomerization of this chromophore. The photocyclic reaction has
been a subject of intensive studies experimentally and theoretically. Upon
flash excitation of the chromophore, the ground state (pG, lmax=446nm)
is converted into a red-shifted intermediate (pR, lmax=465nm) in less than
2 ns. Subsequently pR decays in a sub-millisecond time scale into a blue
shifted intermediate (pB, lmax=355nm), which returns to pG in a sub-sec
time scale. We studied a time-resolved energies and volume changes as well
as the diffusion coefficients of intermediate species during the photocyclic
reaction of photoactive yellow protein (PYP).
@@The traditional spectroscopic techniques are certainly useful and powerful
to characterize the proteins. However, a serious limitation inherent in
the traditional techniques is that they are applicable only to steady state
protein structures. Knowledge of these properties of time-dependent or
unstable (intermediate) species during biological reactions is very limited,
which prevents us from using the compiled data to characterize the intermediate
structures of proteins. It is most desirable to develop and use a method
that can measure these properties in the time domain so that reaction intermediates
can be characterized in a similar way.
@@Analyzing the TG signal in the 1 ~ 20 ms time scale, we determined that
the volume change (delV) for pG->pR is negative and the absolute value
of delV increases with decreasing the temperature. We further analyzed
the TG signal from a few 100 ns to tens milliseconds. Together, the enthalpy
change to the second intermediate is also estimated using the transient
lens (TrL) method. Furthermore, we 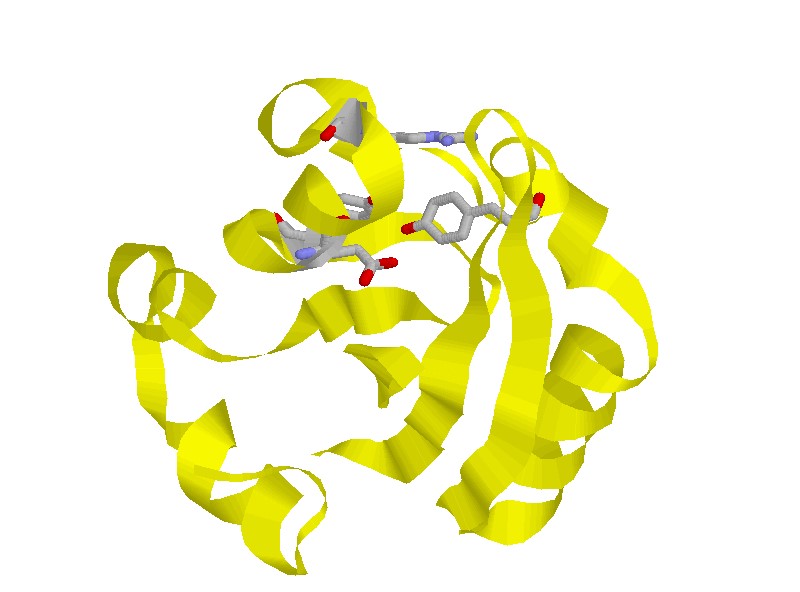 found that the D value of pB is about 0.8 times larger than that of the pG state. By measuring D of PYP denatured by guanidinium hydrochloride, the smaller D is interpreted in terms of the unfolded nature of pB, and the extent of the unfolding in the pB state is estimated. The temperature dependent delV is interpreted in terms of the larger thermal expansion coefficient of pR compared with that of pG. Based on the compiled data on the partial molar volume of native and unfolded states of proteins so far obtained, we suggest that all of these data indicate the partially unfolded nature of pR as well as pB.
found that the D value of pB is about 0.8 times larger than that of the pG state. By measuring D of PYP denatured by guanidinium hydrochloride, the smaller D is interpreted in terms of the unfolded nature of pB, and the extent of the unfolding in the pB state is estimated. The temperature dependent delV is interpreted in terms of the larger thermal expansion coefficient of pR compared with that of pG. Based on the compiled data on the partial molar volume of native and unfolded states of proteins so far obtained, we suggest that all of these data indicate the partially unfolded nature of pR as well as pB.
@@We investigated the structural dynamics as well as the enthalpy changes
of some site-directed mutants of PYP in order to obtain further information
of the dynamics and the structures. If the structure change in pR is not
restricted around the chromophore and the whole protein structure is loosened
as the previous studies suggested, any one residue mutation may not change
the essential features of thermal expansion coefficient change or D as
long as the photocycle reaction takes place.
@@We also used three mutants, R52Q, P68A and W119. Interestingly, on ms
time scale, we observed a new dynamics that has never been detected by
the other spectroscopic methods so far. Since, on this time scale, pR is
already created, this new dynamics indicates that the protein structure
far from the chromophore is still moving after the pR state is created.
Therefore, the pR state is not a single state, but subsequent change of
the protein structure apart from the chromophore is essential to finally
prepare pR leading pB; that is, the protein structure around the chromophore
is initially changed within 3 ns and then the other protein part moves
with a ms lifetime. This dynamics depends on the mutation. This observation
may be the most direct evidence for the global change of the protein in
pR. Based on this result, the two species in pR, of which structures apart
from the chromophore site are different, are called pR1 and pR2.
The diffusion coefficient (D) changes of several N-truncated PYPs, which
lacked the N-terminal 6, 15, or 23 amino acid residues (T6, T15, and T23,
respectively) were investigated. For intact PY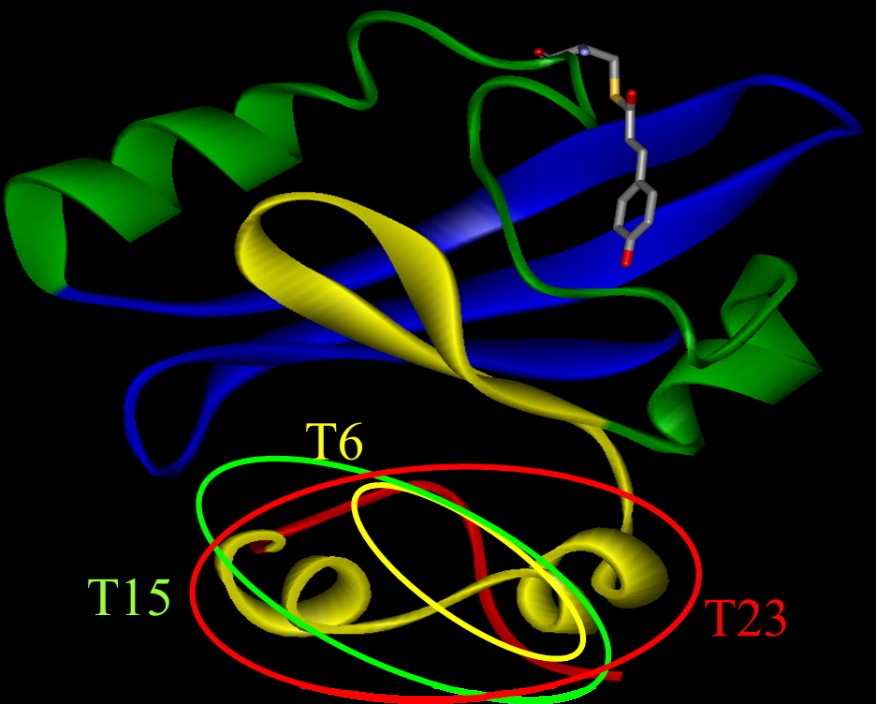 P (i-PYP), D of pB (DpB) was c.a. 11% lower than that (DpG) of the ground
state (pG) species. The difference in D (DpG - DpB) decreased upon cleavage
of the N-terminal region in the order of i-PYP>T6>T15>T23. This
trend clearly showed that conformational change in the N-terminal group
is the main reason for the slower diffusion of pB. This slower diffusion
was interpreted in terms of the unfolding of the two a-helices in the N-terminal
region, increasing the intermolecular interactions due to hydrogen bonding
with water molecules. The increase in friction per one residue by the unfolding
of the a-helix was estimated to be 0.3X10-12 kg/s.
P (i-PYP), D of pB (DpB) was c.a. 11% lower than that (DpG) of the ground
state (pG) species. The difference in D (DpG - DpB) decreased upon cleavage
of the N-terminal region in the order of i-PYP>T6>T15>T23. This
trend clearly showed that conformational change in the N-terminal group
is the main reason for the slower diffusion of pB. This slower diffusion
was interpreted in terms of the unfolding of the two a-helices in the N-terminal
region, increasing the intermolecular interactions due to hydrogen bonding
with water molecules. The increase in friction per one residue by the unfolding
of the a-helix was estimated to be 0.3X10-12 kg/s.
@@@@@@@@@@@@@@@@@@@@@@ (Back)


photo-physical-chemistry lab,sεwεw@w€Θ »wκU υ¨»w€Ί
§606-8502
Kitashirakawaoiwakecho
Sakyoku, Kyoto, Japan
TEL +81-75-753-4026
FAX +81-75-753-4000
Links for members
Bake Web mail (Set up)@
Manuals